SEARCH
Search
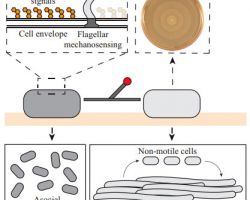
Motility
-
General
Motility
Bacteria use different motility patterns to navigate and explore natural habitats.
- Swarming
- Swimming
- Twitching
- Gliding
- Sliding
- Brownion
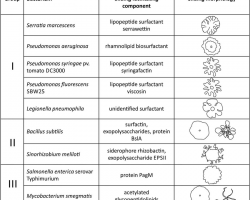
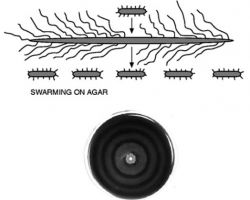
♦ Swarming
Is a rapid (2–10 μm/s) and coordinated translocation of a bacterial population across solid or semi-solid surfaces, and is an example of bacterial multicellularit and swarm behaviour.
Swarming motility has been mostly studied in genus Serratia, Salmonella, Aeromonas, Bacillus, Yersinia, Pseudomonas, Proteus, Vibrio and Escherichia.
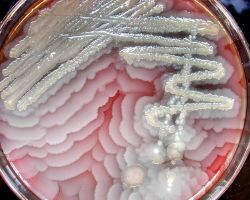
♦♦ Bacillus subtilus
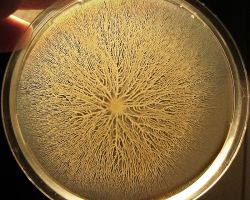
Bacteria of the species Bacillus subtilis were inoculated at the center of a dish with gelose containing nutrients.
The bacteria start mass-migrating outwards about twelve hours after inoculation, forming dendrites which reach the border of the dish (diameter 90mm) within a few hours. Two days after inoculation the number of bacteria has increased so much that they diffuse light significantly and appear white.
This multicellular behavior has been mostly observed in controlled laboratory conditions and relies on two critical elements:
1) the nutrient composition
2) viscosity of culture medium (i.e. % agar).
One particular feature of this type of motility is the formation of dendritic fractal-like patterns formed by migrating swarms moving away from an initial location. Although the majority of species can produce tendrils when swarming, some species like Proteus mirabilis do form concentric circles motif instead of dendritic patterns.
♦♦ Proteus mirabilis morphology and swimming on agar
Short swimmer cells, when plated on an agar surface differentiate into elongated, multinuclear heavily flagellated swarmer cells (up to 80 μm long).
Swarmer cells associate with one another and spread away from the point of inoculation. After several hours, swarmer cells consolidate into numerous swimmer cells thus completing the cycle.
Alternating cycles of differentiation and consolidation result in a bull's-eye pattern of swarming on an agar plate.
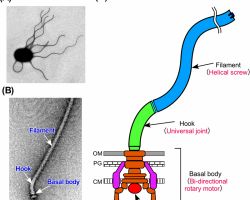
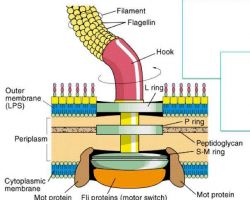
♦ Flagella: structure, types and function
- Flagellum (singular) is hair like helical structure emerges from cell wall and cell membrane
- It is responsible for motility of the bacteria
- Size: thin 15-20nm in diameter.
♦♦ Structure
- Flagella is not straight but is helical
- It is composed of flagellin protein (globular protein) and known as H antigen.
-The flagellum consists of at least three part: the basal body as a bi-directional rotary motor, the hook as a universal joint and the filament as a helical screw.
♦♦ Flagellar arangement
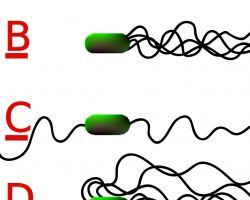
Different species of bacteria have different numbers and arrangements of flagella.
- Monotrichous bacteria have a single flagellum (e.g., Vibrio cholerae)
- Lophotrichous bacteria have multiple flagella located at the same spot on the bacterial surfaces which act in concert to drive the bacteria in a single direction.
- Amphitrichous bacteria have a single flagellum on each of two opposite ends (only one flagellum operates at a time, allowing the bacterium to reverse course rapidly by switching which flagellum is active).
- Peritrichous bacteria have flagella projecting in all directions (e.g., E. coli).
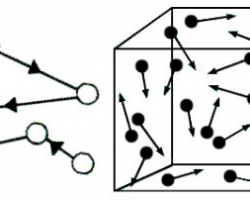
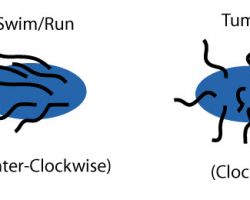
Counterclockwise rotation of a monotrichous polar flagellum pushes the cell forward with the flagellum trailing behind, much like a corkscrew moving inside cork.
Flagella are left-handed helices, and bundle and rotate together only when rotating counterclockwise. When some of the rotors reverse direction, the flagella unwind and the cell starts "tumbling".
Even if all flagella would rotate clockwise, they likely will not form a bundle.
Such "tumbling" may happen occasionally, leading to the cell seemingly thrashing about in place, resulting in the reorientation of the cell. The clockwise rotation of a flagellum is suppressed by chemical compounds favorable to the cell (e.g. food), but the motor is highly adaptive to this. Therefore, when moving in a favorable direction, the concentration of the chemical attractant increases and "tumbles" are continually suppressed; however, when the cell's direction of motion is unfavorable (e.g., away from a chemical attractant), tumbles are no longer suppressed and occur much more often, with the chance that the cell will be thus reoriented in the correct direction.
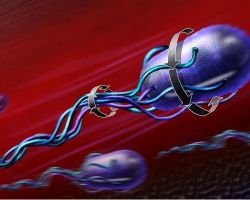
♦ Swimming
Bacterial cells like Escherichia.coli swim by rotating a 'propeller' made from a bundle of long thin filaments. The bacterial flagellar motor is the tiny nanomachine that rotates these filaments.
When all the motors at the base of each filament rotate counter-clockwise together, the filaments bundle to form a propeller that rotates counter-clockwise, and the bacterium swims through its environment rotating subtly the other way while it moves to where life is better.
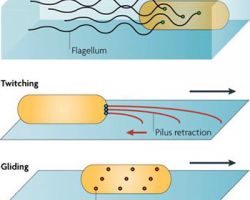
♦ Twitching / crawling
Is a form of crawling bacterial motility used to move over surfaces. Twitching is mediated by the activity of hair-like filaments called type IV pili which extend from the cell's exterior, bind to surrounding solid substrates and retract, pulling the cell forwards in a manner similar to the action ofa grappling hook.
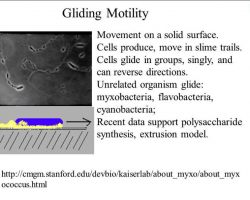
♦ Gliding
Bacterial gliding is a process of motility whereby a bacterium can move under its own power. This process does not involve the use of flagella, pili, or fimbriae, all of which are more well-understood apparatuses used in bacterial motility. The mechanisms of gliding are only partially known. Generally, the process occurs whereby the bacterium moves along a surface in the general direction of its long axis. Gliding may occur via distinctly different mechanisms, depending on the type of bacterium. This type of movement has been observed in phylogenetically diverse bacteria, such as cyanobacteria, myxobacteria, cytophaga and mycoplasmas, and may play an important role in biofilm formation, bacterial virulence and chemosensing.
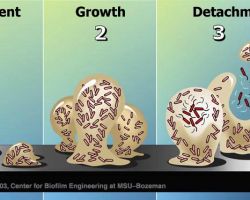
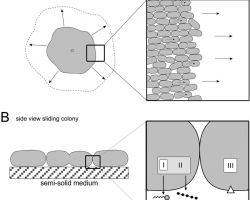
♦ Sliding
Mechanism of sliding motility, the expansion powered by the pushing force of dividing cells.
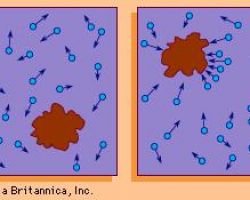
♦ Brownian
Brownian motion is the random, uncontrolled movement of particles in a fluid as they constantly collide with other molecules
They are in part responsible for facilitating movement in bacteria that do not encode or express motility appendages, such as Streptococcus and Klebsiella species.
Brownian motion can also affect “deliberate” movement exhibited by inherently motile bacteria that harbor pili or flagella. For example, an Escherichia coli cell that is swimming toward an area of higher oxygen concentration may fall “off-track” if it physically encounters a particle moving by Brownian motion or if such a particle(s) obstructs the bacterial cell’s path of motion. This form of “interference” adds to the stochasticity with which bacterial direction can change.
♦♦♦ CLICK ON PHOTO BROWNIAN MOTION ♦♦♦
- This is a simulation of the Brownian motion of 5 particles (yellow) that collide with a large set of 800 particles. The yellow particles leave 5 blue trails of random motion and one of them has a red velocity vector.
- This is a simulation of Brownian motion of a big particle (dust particle) that collides with a large set of smaller particles (molecules of gas) wich move with different velocities in different random direction.
-
History
-
Related
-
References
wikipedia
fotoos
wikipedia
https://www.biorxiv.org/content/biorxiv/early/2018/08/23/398321.full.pdf
https://www.sciencedirect.com/topics/immunology-and-microbiology/proteus-mirabilis
https://phys.org/news/2017-08-nanomachines-bacteria.html
http://www.onlinebiologynotes.com/bacterial-flagella-structure-types-function/+
https://www.mit.edu/~kardar/teaching/projects/chemotaxis(AndreaSchmidt)/finding_food.htm
https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology
https://www.researchgate.net/figure/Mechanism-of-sliding-motility-A-Top-view-of-an-expanding-sliding-colony-left-On-the_fig1_315747793



-250x200.jpg)